The discovery of antibodies that foil almost every HIV variant has transformed the AIDS vaccine search.
In 2006, the International AIDS Vaccine Initiative (IAVI) launched a worldwide hunt to find the rarest of immune system warriors against HIV—antibodies that could thwart almost every known strain of the virus. IAVI, a New York City–based nonprofit, aptly gave the project a Mission Impossible–sounding moniker: Protocol G. If it succeeded, Protocol G promised to invigorate the failure-plagued HIV vaccine field and just possibly help design a product that could end the AIDS epidemic.
HIV at that point had thoroughly humbled vaccine researchers, time and again dodging any immune attack they could devise. More than 2 decades of effort had yet to bring a vaccine to market. But researchers had already glimpsed a weapon they hoped might turn the battle: so-called broadly neutralizing antibodies (bNAbs). Only a handful had been discovered, and even the best of them worked against only a few dozen of the hundreds of HIV strains that a vaccine would have to outwit to make a dent in the global epidemic.

Dennis Burton of Scripps holds a model of an HIV bNAb he bagged.
CREDIT: PHOTO BY DAVE FREEMAN, COURTESY OF THE SCRIPPS RESEARCH INSTITUTE
The Protocol G team made an all-out effort to find more. Researchers analyzed 1800 blood samples taken from HIV-infected people in Zambia, South Africa, India, Thailand, Nigeria, Côte d’Ivoire, Rwanda, Uganda, Kenya, Australia, the United Kingdom, and the United States. After 2 years of combing through the sera, an effort led by IAVI team member Dennis Burton, an immunologist at the Scripps Research Institute in San Diego, California, homed in on one woman. By individually probing 30,000 of the woman’s B cells, the immune actors that produce antibodies, they isolated two remarkable antibodies from her serum.
The two Protocol G antibodies jumped out because they “neutralized,” or rendered incapable of establishing an infection, more than 70% of 162 divergent HIV strains, even at very low doses, Burton and co-workers explained in the 9 October 2009 issue of Science. Suddenly, creating a universal HIV vaccine seemed like more than just a wild dream. “It dawned on us that our little world was never going to be the same,” Burton says.
Their little world had indeed turned upside down, and it had big world consequences. In short order, bNAbs became one of the hottest niches not just in HIV research but in immunology itself. Since 2009, researchers have identified more than 50 new HIV bNAbs, several of which look far better than those two from Protocol G.
Dozens of labs are now trying to unravel the intricate pas de deux between HIV and the immune system that leads it to produce bNAbs, a process they hope to mimic one day with an immunogen—a harmless version of the virus used in a vaccine. And the activity in the HIV field has triggered something of a gold rush to find bNAbs for other diseases, including influenza (see sidebar, p. 1171), hepatitis C, dengue, and West Nile viruses. But researchers have no illusions about the challenge of coaxing the immune system to produce a type of antibody that it naturally generates only rarely and in tiny quantities. “If somebody came up with a real immunogen that would reproducibly elicit a broadly protective response, that would be fabulous,” says David Baltimore, a Nobel Prize–winning virologist whose own group at the California Institute of Technology in Pasadena is attempting to genetically engineer stem cells to make bNAbs. “But I see a very difficult road.”
Inauspicious start
Shortly after investigators proved in 1984 that HIV causes AIDS, the key obstacle to making a vaccine emerged. HIV reproduces at a breakneck speed, sloppily copying its genetic code each time. Many of the resultant mutants thrive, and the variants display different protein antigens. Antibodies, which normally target a single antigen with exquisite specificity, simply can’t keep up with the ever-changing virus. As a result, neither the natural immune response nor the response induced by a conventional vaccine is effective.
Contrast this with viruses that vaccinemakers conquered. Measles virus has so little antigenic variation that an antibody response against one virus works against every relative. Even polio, an RNA virus like HIV that mutates at the same rate, can be derailed with a combination of antigens from three distinct strains of the virus. Unlike HIV, these viruses pay a steep fitness price when their antigens change, making it difficult for them to dodge antibody attacks.
CREDIT: D. CORTI AND A. LANZAVECCHIA, ANNU. REV. IMMUNOL. 31 (2013)
In 1990, researchers from Repligen, a Massachusetts biotech, electrified the AIDS vaccine field with a report in Science of the first bNAb against HIV, far more powerful that any antibody seen to date. More astonishing still, they described the exact viral component, or epitope, that stimulated the production of the antibody in animal experiments. A mere six amino acids at the tip of HIV’s surface protein, gp120, taught the immune system how to stop diverse strains of the virus, they reported.
It sounded too good to be true. It was.
As it turned out, this bNAb was a laboratory artifact that worked only against strains of the virus grown in culture dishes, and it had no impact on “primary” isolates taken directly from infected people. That 1993 discovery (Science, 12 November, p. 980) scuttled hopes about the Repligen product. But the next year, Burton and colleagues at Scripps showed that by aspirating bone marrow from an HIV-infected man and then fishing out antibody genes, they had isolated a bNAb that worked against primary isolates.
To establish an infection, HIV first slips gp120 into a CD4 receptor on the surface of white blood cells. Some antibodies can bind to gp120 and neutralize it such that the virus can’t enter the CD4 cell. But as HIV mutates, gp120 changes shape, evading antibodies. Burton’s bNAb attached to a “conserved” part of gp120 that doesn’t tolerate mutations because it is required by many variants to bind CD4 receptors. This bNAb crippled 17 of 24 primary isolates of the virus at what then seemed liked fantastically low doses (Science, 11 November 1994, p. 1024).
At roughly the same time in Austria, another group discovered bNAbs that worked on a different part of HIV. A team led by Hermann Katinger at the Institute of Applied Microbiology in Vienna identified two bNAbs that acted on gp41, a protein that spans the viral membrane and attaches to gp120, and were capable of neutralizing many HIV isolates. This finding suggested that the virus has more than one Achilles’ heel.
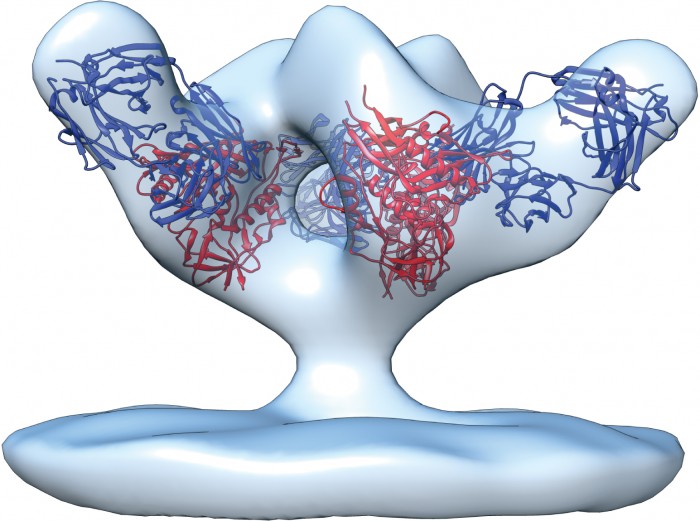
Over the next decade, structural biologists like Ian Wilson at Scripps and Peter Kwong at the U.S. National Institute of Allergy and Infectious Diseases (NIAID) in Bethesda, Maryland, created high-resolution depictions of how these bNAbs bound to both gp120 and gp41. These exquisitely detailed images enabled the investigators to determine the precise mechanisms by which these antibodies tripped up the virus—and why other antibodies had such limited potency and breadth. But few other HIV bNAbs surfaced, and no one found antigens that could trigger their production in animal models or humans. At the same time, antibodies fell out of fashion with many vaccine researchers, who put more stock in stimulating killer cells, a kind of T lymphocyte. While antibodies prevent infection, killer cells identify and destroy cells that the virus manages to enter. Some groups contended that a T cell vaccine alone could stop the virus; others put their stock in a combination T cell/antibody vaccine. Still others rejected the notion of rationally designing a vaccine, pointing to a long history of vaccinologists who succeeded simply by testing various formulations in animals and people, paying little heed to immune mechanisms.
Working backward
Then, two developments reinvigorated the bNAb field.
First, high-profile, T cell–based vaccines against HIV failed miserably (Science, 10 September 2004, p.1545 and 5 October 2007, p. 28), tilting more attention toward antibodies. On the second front, thanks to Protocol G and similar initiatives, the number of bNAbs available for study skyrocketed. Burton and others say that they owe their success to better lures to fish bNAbs from a sea of other antibodies. They also developed improved techniques to clone large numbers of antibodies from single B cells, first amplifying antibody genes and then putting them into viruslike vectors that spit out the products in bulk. For the first time, they could separate these faint immunologic signals from the noise. “Single cell antibody cloning changed the field,” says immunologist Michel Nussenzweig, whose lab at Rockefeller University in New York City helped pioneer the technique for HIV bNAbs (see Review, p. 1199). “Otherwise you didn’t even know what was in the human repertoires.” An extraordinary level of multi-institution collaboration has given the endeavor a major boost, too, through IAVI’s Neutralizing Antibody Consortium and two Centers for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) created by NIAID.
With these advances, researchers can now find bNAbs, albeit at exceedingly low levels, in up to 25% of HIV-infected people. The bNAbs offer scant help to the already infected, as they come too late, when the virus has already infiltrated many blood cells and tissues, and in quantities too small to make a difference. But a vaccine that elicited bNAbs at the right moment, in sufficient quantities, might foil an infection. So researchers are now asking how the immune system makes these antibodies and what gives them their unusual powers.
Like other antibodies, HIV bNAbs emerge as the B cells first exposed to the virus multiply to produce clones that steadily develop more “affinity” for the invader. Called “affinity maturation,” this process of mutation and selection ultimately favors the fittest immune response. But unlike other antibodies, bNAbs appear to evolve slowly over years, accumulating an unusually high number of mutations—three times as many as run-of-the-mill antibodies—which gives them their potency and breadth. The process appears to involve stimulation from different epitopes at different points in time. “There’s an arms race between HIV and the immune system,” Nussenzweig says. “As antibodies chase the virus around, it keeps changing, and the antibodies have to make more mutations.”
Most mutations that give bNAbs their powers take place at the tips of the Y-shaped antibody molecules, which have loops that lasso viral epitopes. Among other things, bNAbs have evolved to become stickier than other antibodies, Nussenzweig and colleagues explained in the 30 September 2010 issue ofNature. That appears to be a response to one of HIV’s unusual properties. The gp120 molecules studding HIV’s surface form clusters of three called trimers. Each HIV has only about 10 trimers on its surface; on influenza viruses, in contrast, the surface proteins form about 450 trimeric spikes. “HIV is almost bald,” Nussenzweig says. This dearth of trimers hampers antibodies: They bind most tightly to a virus when each arm of the Y can separately grab a different trimer. But the extra-sticky bNAbs can compensate by latching on to lipids that make up the HIV viral membrane or even to sugars that coat gp120.
Other peculiar features help HIV bNAbs stymie the virus. As Burton has shown, the loops at the tips of antibody arms are often extra long, increasing their ability to lasso an epitope. Nussenzweig discovered a second, related quirk. Antibody arms typically have a region that remains constant to provide a sturdy framework for the highly mutable loops. But Nussenzweig’s group reported in the 28 March issue ofCell that HIV bNAbs shun convention, accumulating many mutations in the framework region that increase breadth and potency. For unknown reasons, these malleable framework regions don’t compromise the stability an antibody needs to function. “It goes totally against the norm,” Nussenzweig says. “Destabilizing something and making it better is counterintuitive.”
Creating a vaccine that will elicit these features is a daunting assignment, say some researchers. “We’re going to have to make this vaccine with nature’s pathways and guidance,” says Barton Haynes, an immunologist at Duke University in Durham, North Carolina, who heads a CHAVI-ID. “And it’s a vaccine the immune system doesn’t favor making.”
Nussenzweig is optimistic. “People don’t invent vaccines de novo,” he says. “In every case, you take a lesson from nature and try to copy it.” Granted, people with HIV make potent bNAbs in only small amounts, but the discovery that dozens of different ones exist, each of which came about independently, provides hard proof that the human immune system knows how to make them. “If it can happen once,” Nussenzweig says, “it can happen again.”
The making of a bNAb
In 2006, a man in Malawi came to a clinic for sexually transmitted diseases that had a research project under way to try to find people within weeks of becoming infected with HIV. He met the criteria and joined the “acute infection” study, repeatedly donating blood, which enabled researchers to document changes over time in the gp120 of his virus, his antibody response, and ultimately the emergence of a bNAb. Now, this window into antibody evolution is inspiring a strategy to design a bNAb-based vaccine.
A large collaboration led by Duke’s Haynes and John Mascola from NIAID’s Vaccine Research Center studied the man’s sequential blood samples. As the researchers explained in the 25 April issue ofNature, they traced the evolution of the antibody from the initial B cell response all the way to the bNAb, which emerged after more than a year. The original, or germline, antibody did not have much breadth, but it did bind avidly to the original “founder” virus that infected the man. A vaccine, they reason, should use gp120 from the founder virus to kickstart a B cell lineage that can ultimately produce a bNAb. Booster shots could then attempt to guide the mutation of the antibody genes in the cell to expand their breadth by using the same sequence of gp120 variants that naturally created the bNAb in the Malawian man.
In essence, the strategy amounts to carefully selecting a puppy from a litter and giving it the intensive training needed to one day win the best of show. “It’s ingenious that you’d prime with one epitope and boost with another epitope to chaperone the B cell response until the point that it’s truly making a broadly neutralizing antibody,” says immunologist Anthony Fauci, NIAID’s director.
Burton, working with Scripps immunologist William Schief, also tracked the emergence of a bNAb, but they did so retrospectively. They started with a powerful bNAb and used genetic modeling to predict its ancestor’s characteristics. Guided by what they learned, they hope to design a single immunogen that both tickles the right B cells into action and steers them to move from the germline antibody toward the mature bNAb. Unlike the Haynes group, Burton and Schief ‘s team does not want to hold the immune system’s hand and guide the germline B cell to make the bNAb. “Taking that too literally is likely to be asking for too much micromanagement,” Schief says.
As the group explained in the 10 May issue of Science, a screen of massive gp120 libraries led to one that strongly bound both their putative germline antibody and the mature bNAb that evolved from it. The investigators then groomed this gp120 to contain little more than the epitope that both antibodies target. They now hope to show that they can elicit a bNAb response by “priming” the immune system with this artificial gp120, then following with a booster that contains trimer spikes in the most natural configuration possible.
Both the Burton/Schief team and Haynes’s group have monkey studies under way to test their ideas. But even if they learn how to generate bNAbs, Fauci notes that a fundamental, proof-of-principle experiment has yet to happen: No one has shown that giving bNAbs to a person can prevent an infection. To that end, the Vaccine Research Center at NIAID hopes to soon launch human studies that passively give monoclonal bNAbs—which theoretically could last in the body a few months—to newborns of HIV-infected mothers and, separately, to adults at high risk of becoming infected. “Whether they’re going to prevent acquisition of HIV is an open question,” Fauci cautions.
Even if the bNAb strategy passes all these hurdles, years of testing will still be needed to prove that the vaccine protects people and is safe. But Fauci and many others at the forefront of the HIV vaccine search say bNAbs have brought a level of hope and common purpose that this fractious, battle-weary field has rarely, if ever, enjoyed.